This is an unofficial archive of PsychonautWiki as of 2025-08-11T15:14:44Z. Content on this page may be outdated, incomplete, or inaccurate. Please refer to the original page for the most up-to-date information.
UserWiki:Meisam: Difference between revisions
Jump to navigation
Jump to search
>Meisam |
>Meisam m →FAQ |
||
| Line 30: | Line 30: | ||
*:RMS Displacement: 0.001200 | *:RMS Displacement: 0.001200 | ||
*Why I am not using more accurate method (larger basis set, other XC-functionals, solvents effect, multireference methods, etc.) for my calculations? | *Why I am not using more accurate method (larger basis set, other XC-functionals, solvents effect, multireference methods, etc.) for my calculations? | ||
*: | *:More accurate calculations need lots of extra computation power which currently I don't have. | ||
==My works== | ==My works== | ||
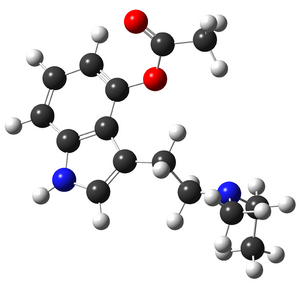
[[File:4AcoMET-UAM1.png|thumb|[[4-AcO-MET]]]] | [[File:4AcoMET-UAM1.png|thumb|[[4-AcO-MET]]]] | ||
Revision as of 20:47, 7 September 2016
About me
Theoretical molecular chemist/physicist. Interested in finding stable geometries of molecules (mostly research chemicals) as a base for further research by other scientists.
FAQ
- What do I do here?
- I calculate the stable geometry of the molecules with Austin Model 1, (AM1) and B3LYP density functional approximation method and upload my results here.
- How accurate are these results?
- It is hard to say. But AM1 results are fairly good and B3LYP results are mostly accurate for geometry of organic molecules.
- Why the molecular bonds/shapes look different than the skeletal formula representation?
- Skeletal formula predictions for bonds/shapes are not accurate. I'm calculating the geometry which have the least energy (therefore is the most stable one). These geometries may or may not be the same shape as their skeletal formula. The visualization software that I use, is guessing the bond-type based on the bond length and atomic type. (which may be a better estimation!) The correct way to find the bond order is to perform a NBO analysis to accurately determine the bond types. (Unfortunately, I don't have enough free time to do that)
- Are these geometries valid for molecules in water/SBF/blood/urine, etc?
- No! All my calculations are done on isolated molecules (basically molecules in low pressure gas phase). The geometries depend on the environment and do change in presence of solvents. But based on the property you are interested in, they are generally a good approximation or a first guess.
- Are these geometries the most stable molecular configuration?
- There's no guarantee for that. I'm just searching for local energy minima on potential energy surface. These molecules have a high degree of freedom and I have not enough time to check for all stable geometries and find the most stable one.
- I start my B3LYP calculations from the geometry obtained by AM1 method and usually do not check for imaginary vibrational frequencies.
- Can people use these geometries for their own research?
- Absolutely! Please read the terms of use for CC-BY-SA-4.0 license.
- Also keep in mind that B3LYP implementation of Gaussian (VWN functional part) may be different from the one you are going to use in other computational packages.
- What is the Z-matrix?
- A method for representing molecular geometries. You can use that for you research or to make a better image of molecules (if you want to!) More info here!
- Which software do I use?
- I use Gaussian 09 for calculations and GaussView 5 for visualization.
- Which basis set I am using?
- Unless stated otherwise:
- Polarized split-valence Pople-style Gaussian basis set 6-31G**
- What are my convergence criteria?
- Unless stated otherwise:
- Maximum Force: 0.000450
- RMS Force: 0.000300
- Maximum Displacement: 0.001800
- RMS Displacement: 0.001200
- Why I am not using more accurate method (larger basis set, other XC-functionals, solvents effect, multireference methods, etc.) for my calculations?
- More accurate calculations need lots of extra computation power which currently I don't have.
My works